Clinical characteristics of the recovered COVID-19 patients with
re-detectable positive RNA test
Authors: Jianghong An,
1,*
Xuejiao Liao,
2,*
Tongyang Xiao,
2,*
Shen Qian,
2,*
Jing Yuan,
3
Haocheng Ye,
2
Furong Qi,
2
Chengguang Shen,
2
Yang Liu,
2
Lifei Wang,
4
Xiaoya
Cheng,
1
Na Li,
2
Qingxian Cai,
5
Fang Wang,
5
Jun Chen,
5
Yingxia Liu,
3
Yunfang Wang,
6
Feng Zhang,
7
Yang Fu,
8
Xiaohua Tan,
1,†
Lei Liu,
2,9,†
Zheng Zhang
2,9,†
Author Affiliations:
1
Department of Oncology and Hematology, Shenzhen Third People’s Hospital, Shenzhen
518112, Guangdong Province, China
2
Institute of Hepatology, National Clinical Research Center for Infectious Disease,
Shenzhen Third People’s Hospital, Shenzhen 518112, Guangdong Province, China
3
Department of Infectious Diseases, Shenzhen Third People’s Hospital, Shenzhen
518112, Guangdong Province, China
4
Department of Radiology, Shenzhen Third People’s Hospital, Shenzhen 518112,
Guangdong Province, China
5
Department of Hepatology, Shenzhen Third People’s Hospital, Shenzhen 518112,
Guangdong Province, China
6
Translational Research Center, Beijing Tsinghua Changgung Hospital, Tsinghua
University, Beijing 102218, Beijing , China
7
Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge,
MA 02139, USA
8
School of Medicine, Southern University of Science and Technology, Shenzhen,
Guangdong, 518055, China
9
The Second Affiliated Hospital, School of Medicine, Southern University of Science and
Technology, Shenzhen 518112, Guangdong Province, China
*Contributed equally
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint
NOTE: This preprint reports new research that has not been certified by peer review and should not be used to guide clinical practice.

†Joint corresponding authors
Corresponding Authors:
Prof. Zheng Zhang, PhD, MD. Institute of Hepatology, Shenzhen 3rd People’s Hospital,
Shenzhen 518112, Guangdong Province, China; Phone: 86-755-81238983; Fax:
86-755-81238983; Email: zhangzheng1975@aliyun.com.
Prof. Lei Liu, MD, Shenzhen 3rd People’s Hospital, Shenzhen 518112, Guangdong
Province, China; Email: liulei332[email protected]om.
Prof. Xiaohua Tan, PhD, MD. Department of Oncology and Hematology, Shenzhen 3rd
People’s Hospital, Shenzhen 518112, Guangdong Province, China; Email:
xiaohua_t@hotmail.com.
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint
Summary
Background It has been reported that several cases recovered from
COVID-19 tested positive for SARS-CoV-2 after discharge (re-detectable
positive, RP), however the clinical characteristics, significance and potential
cause of RP patients remained elusive.
Methods A total of 262 COVID-19 patients were discharged from January 23
to February 25, 2020, and were enrolled for analysis of their clinical
parameters. The RP and non-RP (NRP) patients were grouped according to
the disease severity during their hospitalization period. The clinical
characterization at re-admission to the hospital was analyzed. SARS-CoV-2
RNA and plasma antibody levels were detected using high-sensitive detection
methods.
Findings Up to March 10, 2020, all of patients were followed up for at least 14
days, and 38/262 of RP patients (14.5%) were present. The RP patients were
characterized by being less than 14-years old and having mild and moderate
conditions as compared to NRP patients, while no severe patients became RP.
Retrospectively, the RP patients displayed fewer symptoms, more sustained
remission of CT imaging and earlier RNA negative-conversion but similar
plasma antibody levels during their hospitalization period as compared to
those NRP patients. When re-admitted to the hospital, these RP patients
showed no obvious clinical symptoms or disease progression indicated by
normal or improving CT imaging and inflammatory cytokine levels. All 21 close
contacts of RP patients were tested negative for SARS-CoV-2 RNA, and no
suspicious clinical symptoms were reported. However, 18/24 of RNA-negative
samples detected by the commercial kit were tested to be positive for virus
RNA using a hyper-sensitive method, suggesting the carrier status of virus
possibly existed in patients recovered from COVID-19.
Interpretation Our results showed that young and mild COVID-19 patients
seem to be RP patients after discharge, who show no obviously clinical
symptoms and disease progression upon re-admission. More sensitive RNA
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint
detection methods are required to monitor these patients during follow-up. Our
findings provide empirical information and evidence for the effective
management of COVID-19 patients during their convalescent phase.
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint
Introduction
Since early December of 2019 and up to March 23, 2020, over 300, 000 cases
of coronavirus disease 2019 (COVID-19) caused by novel coronavirus
(SARS-CoV-2) infection, with over 13, 000 deaths have been reported through
the world [1]. The World Health Organization has declared COVID-19 as a
pandemic [2]. Generally, the COVID-19 is less severe and less fatal than the
SARS, however, some patients, especially those who are elderly with
co-morbidities are prone to develop more severe symptoms and require
emergent medical interventions [3, 4]. Many literatures have retrospectively
analyzed the clinical characteristics of patients infected with SARS-CoV-2 [3-8].
Recently, an increasing number of patients with COVID-19 were discharged
from the hospital and received regular follow-up and observation.
Re-detectable positive (RP) of SARS-CoV-2 RNA test in some recovered
patients has been reported [9-12]. The management of RP patients has
attracted wide attention. However, the number of RP patients reported in the
literature was small, and the duration of follow-up was short. In addition, the
clinical characteristics is lacking and the potential impact and significance of
RP patients remain unknown, which makes it difficult to provide empirical
information and evidence support for the management of patients with
COVID-19 in the recovery period.
This study retrospectively analyzed the clinical characteristics of 38 RP
patients and 224 non-RP (NRP) patients recovered from COVID-19. It is found
that RP patients were characterized by younger age and milder conditions.
They also had minor symptoms, more sustained remission of CT imaging and
earlier RNA negative-conversion but similar plasma antibody levels during their
hospitalization period. They showed no obvious disease progression and
infectivity when re-admitted to the hospital. The hyper-sensitive detection
method identified SARS-CoV-2 RNA molecules from most samples that were
tested RNA-negative by the commercial kit, suggesting the carrier status of
virus possibly existed in recovering COVID-19 patients. These findings provide
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint
key information for the effective management of COVID-19 patients during
their convalescent phase.
Methods
Study design and participants
A total of 262 confirmed COVID-19 patients discharged from Shenzhen Third
People's Hospital from January 23, 2020 to February 25, 2020 were enrolled in
this study. All discharged COVID-19 patients were continued to be isolated and
observed for 14 days, weekly followed-up and SARS-CoV-2 RNA detection
were performed timely. All discharged patients were followed-up for at least
two additional weeks after isolation. Among them, the RP patients were
re-admitted to hospital for further medical observation and close contacts were
also followed-up. The rest of the recovered NRP patients were closely
followed-up outside the hospital. This study was approved by the Ethics
Committee of The Third People's Hospital of Shenzhen (2020-115), which
waived the requirement for written patient consent for this retrospective
analysis. All patients gave their oral consent to participate in this retrospective
study.
Clinical definition
According to the guideline of the diagnosis and treatment for novel coronavirus
pneumonia (the sixth edition) published by National Health Commission of the
People’s Republic of China [13], all first diagnosis cases of COVID-19 were
confirmed according to positive respiratory RT-PCR tests. The discharge
criteria of the recovered patients included: temperature returned to normal for
more than 3 days, respiratory symptoms significantly improved, and significant
absorption of pulmonary lesions of chest CT imaging, and at least consecutive
negative RNA test results for 2 apart from each other by at least 24 hours. The
RP patients were confirmed by digestive (anal swab) and respiratory positive
RT-PCR tests. Since February 22, 2020, evaluation of negative anal swab was
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint
supplemented for discharge criteria in Shenzhen Third People's Hospital.
Data collection
The medical records of 262 recovered COVID 19 patients including 38 RP
patients were reviewed. The epidemiological, demographic, clinical, laboratory
data of the patients were collected, summarized and analyzed. According to
the first chest CT imaging post admission, the extent of pulmonary
inflammation was divided into mild, moderate and severe condition basing on
the lesions involving unilateral lobe, multiple lobes in both lungs, and all lobes
in both lungs, respectively. According to chest CT within 7 days after admission,
the remission of the lesions was evaluated. The temporary progression was
indicated by increased lesion and persistent remission was indicated by stable
or absorbed or decreased lesions.
qRT-PCR and Sherlock assay for SARS-CoV-2 RNA detection
The quantitative reverse transcription polymerase chain reaction (qRT-PCR)
was assessed as described previously [14]. Nasopharyngeal and anal
specimens collected during hospitalization were sent to the laboratory in viral
transport case. Total nucleic acid extraction were extracted from the samples
using the QIAamp RNA Viral Kit (Qiagen, Heiden, Germany), and quantitative
RT-PCR was performed using a China Food and Drug Administration (CFDA)
approved commercial kit specific for 2019-nCoV detection (GeneoDX Co., Ltd.,
Shanghai, China) or Sherlock kit gifted from Feng Zhang lab according to the
manual. Each RT-PCR assay provided a Ct value, which is the number of
cycles required for the fluorescent signal to cross the threshold for a positive
test, a higher Ct value is correlated with a lower viral load. The specimens
were considered positive if the Ct value was ≤ 37.0, and negative if the viral
load were undetectable. Specimens with a cycle-threshold value higher than
37 were repeated. The specimen was considered positive if the repeat results
were the same as the initial result and between 37 and 40. If the repeat Ct was
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint
undetectable, the specimen was considered negative. All procedures involving
clinical specimens and SARS-CoV-2 were performed in a biosafety level 3
laboratory. Meanwhile, we did next-generation sequencing of samples from
three patients.
ELISA assay for anti-SARS-CoV-2 IgG and IgM antibody
Microtiter plates (Sangon Biotech) were coated overnight at 4°C with 4 μg/mL
recombinant SARS-CoV-2-RBD (Receptor binding domain) proteins (50 μL
per well) expressed by our laboratory through 293-T cells. The plates were
washed thrice with PBS containing 0.1% v/v Tween-20 (PBST) and blocked
with blocking solution (PBS containing 2% w/v non-fat dry milk) for 2 hours at
37°C. The plates were then washed with PBST. The sera were diluted to
200-fold into PBS as an initial concentration, and serial 3-fold dilutions of sera
was added to the wells and incubated at 37°C for 60 minutes. After three
washes, 100 μL of horseradish peroxidase (HRP)-conjugated goat anti-human
IgG (for IgG antibody titer detection) and IgM (for IgM antibody titer detection)
antibodies solution (Sangon Biotech) were added to each plate, respectively,
and incubated at 37°C for 60 minutes. After five washes, 100 μL of
tetramethylbenzidine (TMB) substrate (Sangon Biotech) was added at room
temperature in the dark. After 15 minutes, the reaction was stopped with a 2 M
H2SO4 solution. The absorbance was measured at 450 nm. All samples were
run in triplicate. The ELISA titers were determined by endpoint dilution.
Statistical analysis
A statistical analysis was performed using SPSS 26.0 (IBM, Chicago). All of
the statistical tests were two-sided, and significant differences were
considered at p < 0.05. Continuous variables were evaluated using the median
and interquartile range (IQR) values. Chi-square or Fisher exact tests were
utilized to compare the proportions of the categorical variables.
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint
Results
Demographic, epidemiological and clinical characteristics
A total of 262 patients were discharged from January 23, 2020 to February 25,
2020 and were followed-up for at least 14 days. Among them, mild, moderate
and severe patients accounted for 11.4% (n = 30), 81.0% (n = 212) and 7.6%
(n = 20), respectively. Up to March 10, 14.5% of convalescent patients (n = 38)
were re-detected to be SARS-CoV-2 RNA positive during their followed-up
period. None of severe patients were re-tested to be RNA positive
(Supplemental Table 1).
It revealed that the vast majority of RP patients (97.4%, n = 37) were
younger than 60 years of age. Among them, patients younger than 14 years
old were more common compared with those between the ages of 14 and 60
years (35.0% vs 16.0%, p < 0.01) (Table 1). In addition, it is found that 36.7%
(11/38) of RP patients are characterized by mild symptoms. The percentage
was significantly higher than what was seen among NRP patients (12.7%,
19/204, p < 0.01, Supplemental Table 1). There was no significant difference in
the gender distribution. Notably, there were less mild RP patients having fever
in their initial symptoms as compared to mild NRP patients (p < 0.01). Also,
45.5% of mild RP patients displayed only upper respiratory symptoms at the
first admission, while mild NRP patients usually had lower respiratory
symptoms at the first admission (Table 1). There is no difference of the extent
of lesions in the first chest CT imaging between RP and NRP patients with
moderate stages. However, the incidence of RP (85.2%) was found to be
particularly closely related to the sustained remission of chest CT imaging as
compared to NRP patients, of which 36.2% displayed transient progression
during their first hospitalization period (Table 1 and Figure 1). There was no
significant difference in the usage of steroid and antiviral therapy between RP
and NRP patients during their first hospitalization period. In addition, RP
patients did not show a higher incidence of a history of traveling and living in
Hubei province as compared to NRP patients.
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint
Differential RNA dynamic in RP patients with that in NRP patients
No differences of days of last RNA negative-conversion since the onset of
illness and hospitalization days were found between RP and NRP patients.
Importantly, RNA negative-conversion occurred mostly within 2-3 weeks since
the onset of illness among 63.6% mild RP patients and within 1-2 weeks since
the onset of illness among 22.2% moderate RP patients. By contrast, there are
more NRP patients who displayed RNA negative-conversion after 3 weeks
since the onset of illness regardless of mild or moderate status (Table 2).
These data showed RP patients were characterized by early RNA-negative
conversion while NRP patients cleared virus relatively late.
The changes of serum anti-SARS-CoV-2 IgG and IgM antibodies in
patients recovered from COVID-19
In order to evaluate the effect of serum-specific antibody levels on the
occurrence of RP, we analyzed the difference of anti-SARS-CoV-2 IgG and
IgM antibody levels in both RP and NRP patients at their discharge. More than
half of RP and NRP patients displayed medium levels of IgG and IgM
independent on their disease severity. However, there are no differences of
antibody levels in between both groups of patients (Supplemental Table 2). We
also evaluated the dynamic of IgG and IgM levels at the discharge and
re-admission in RP patients. The IgG and IgM levels were maintained at stable
levels in these RP patients during the 14 days period (data not shown).
Supplementing negative results of anal swab test at discharge failed to
reduce RP occurrence of COVID-19 patients
In order to evaluate the effects of increasing sampling site test at discharge on
RP events, we compared the occurrence events of RP before Feb 22 and after
Feb 22 after that time the negative anal swab detection was added to
discharge criterion in COVID-19 patients. Our results showed that there was
no statistical difference in the occurrence of RP patients before Feb 22 and
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint
after Feb 22 (14.5% vs 14.3%, p = 0.77, Figure 2). These data indicated that
supplementing detected sites of SARS-CoV-2 RNA failed to reduce the RP
occurrence in convalescent patients.
RP patients showed no obvious clinical symptoms and disease
progression.
All 38 RP patients were re-admitted to the hospital for further medical
observation. The analysis showed that all these patients had no fever. A small
number of patients reported mild cough and chest tightness, which were not
worse than before (Table 3). All patients recovered from mild conditions (n = 30)
and 37.0% from moderate patients had normal chest CT imaging without
inflammatory signs. By contrast, 63.0% (n = 17) of patients recovered from
moderate conditions had stable or reduced inflammatory signs in their chest
CT imaging (Figure 1). There were normal range of the lymphocyte count,
plasma IL-6 and CRP levels upon admission for all RP patients. Only one
patient received transient interferon-alpha inhalation therapy, and 4 patients
received low-flow oxygen inhalation therapy and traditional Chinese medicine
after admission.
In addition, because all the convalescent patients with COVID-19 in our
cohort were required to be isolated at home or under intensive isolation, only
21 close contacts were produced. Up to March 10, 2020, all of 21 close
contacts were tested to be negative for SARS-CoV-2 RNA, and no suspicious
clinical symptoms were reported in those close contacts.
Hyper-sensitive methods potentially improved SARS-CoV-2 RNA
detection in RP patients
To investigate the possible false negative due to low-sensitive commercial
RNA detection kit, we used a higher-sensitive method to detect various types
of samples from both RP patients and NRP patients with similar illness days.
For 24 samples from 15 of RP patients who were sampled after 5-7 days since
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint
the onset of the re-admission, 75% of spike genes and 41.6% ORF genes
could be detected to be positive using hyper-sensitive method, while only
12.5% N genes and 4.2% RF genes were detected to be positive using
commercial detection kit. Eight of fifteen RP patients were confirmed to be
RNA positive using the hyper-sensitive kit, although only 1 person was
confirmed using commercial kit. By contrasts, 8 samples from NRP patients
were detected to be negative by both methods. These data showed
hyper-sensitive methods potentially improve RNA positive detection in
samples from RP patients with negative results.
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint
Discussion
Several studies have shown the existence of RP patients [9-12], however their
clinical characterization as not well defined. This study retrospective analyzed
the clinical and followed-up data in a cohort of RP and NRP patients in the
same discharge period. Up to March 10, 2020, 38 RP patients were present
which accounts for 14.5% of discharged patients during the same followed-up
period. These RP patients displayed several significant features, including
younger age and mild and/or moderate symptom during their hospitalization,
which is in consistent with a previous report [9, 12]. Mild RP patients were
usually younger than 14 years old and moderate RP patients were younger
than 60 years old. By contrast, no severe patients were found to be RP
patients within the similar follow-up period. In addition, more RP patients
displayed minor symptoms in their hospitalization such as less comorbidities
and fever, and more upper respiratory symptoms. RP patients also maintained
more remission in their CT imaging than those of NRP patients. These data
indicated that RP patients were characterized by younger and minor
symptoms in their hospitalization period.
Virus load is usually thought to be related to the disease outcome [14, 15].
The present study indicated RNA negative-conversion occurred commonly 2-3
weeks since the onset of illness in moderate RP patients as compared to more
than 3 weeks in moderate NRP patients. The significantly shortened RNA
negative-conversion time may affect the persistence of high levels of adaptive
immunity [16]. Our recent studies indicated that a higher titer of antibody in the
plasma was independently associated with disease severity in patients with
COVID-19 [17]. However, RP and NRP patients displayed similar levels of IgG
and IgM in the plasma. Future study should investigate host immune
responses which were usually considered to determine the clinical outcome
especially in virus infection [18, 19].
We also comprehensively characterized the clinical symptoms of RP
patients when they were re-admitted to the hospital. No obvious clinical
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint
evidence of disease progression or recurrence was found in these RP patients
including CT imaging and laboratory tests. And no antibiotics, steroids,
antiviral agents and continuous supplemental oxygenation were required in
these RP patients. The inflammatory response was significantly reduced.
These data indicated that the diseases of RP patients did not progress into
more severe status even if their RNA was detected to be positive for
SARS-CoV-2. More important, these RP patients have not caused new
infections after discharge. And from a recent longitudinal study in SARS-CoV-2
infected rhesus macaques, reinfection could not occur in convalescent
monkeys [20]. Long-term follow-up of these close contacts with RP patients
will warrant the evaluation of possible risk of RP.
The underlying mechanisms underlying RP occurrence remain unclear. The
possible reasons argued by a large number of experts are related to several
virological, immunological and sampling methodological factors. Virologically,
the false negatives [21], viral residual [12], intermittent viral release [12] and
viral distribution [22, 23] are usually considered to be major factors. Our data
support the notion that the false negatives using commercial kit may partially
account for the RP, because the kits had only 30%–50% positive rate of
detection [23, 24]. In 24 of various samples from RP patients, RNA was
detected to be negative for both N gene and ORF1b gene at several days after
their re-admission to the hospital using commercial kit, whose lower limit of
detection (LOD) was relatively high (500 copies/ml). However, using a more
sensitivity Sherlock kit with an LOD of 100 copies/ml [25], 75% of samples
were detected to be positive for S gene and 41.6% for ORF genes, thus
leading to half of positive subjects present within RP patients with undetectable
RNA using commercial kit in their hospitalization. By contrast, among 8
samples from NRP patients, none was detected to be positive using either
Sherlock or commercial kit. However, in a sample confirmed by SRAS-CoV-2
sequencing, the Sherlock tested it as positive (data not shown). Therefore,
future study should improve both the sensitivity and specificity of detection kit,
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint
which would accurately identified clinical samples. Anyway, these data
indicated that false positive detected by current kit may on some extent
account for the occurrence of RP patients.
Another virological factor is that long-term virus residual in gut and other
tissue, similar to SARS [26]. A recent study indicated that SARS-CoV-2 nucleic
acid can persist in the digestive tract and feces for nearly 50 days [27]. Thus,
extending detection time is necessary for the COVID-19 patients when they
were discharged. However, our results show that adding anal swab derived
RNA tested to be negative as discharge criterion did not significantly reduce
the occurrence of RP patients. Thus other factor may be associated with the
RP patients. We could not exclude sampling methodological factors including
differential sampling and operational methods, sample quality, and technician
expertise levels. Nor could we exclude immunological factors including low
mucosal immune responses such as low IgA levels. These factors may take
some uncertain risks leading the occurrence of RP patients [4, 27]. Future
studies should reduce RP occurrence through using hyper-sensitive detection
kit with hyper-specificity, combining detection of multiple samples with more
immune markers.
This study has several limitations. First, this study is a single-center
retrospective study and the duration of follow-up is short, and more clinical
observations are needed to evaluate the potential risk of SARS-CoV-2
recurrence and infection. Second, dynamics of SARS-CoV-2 RNA in
COVID-19 patients need to be monitored and evaluated for RP patients.
Second, additional studies should measure the dynamic changes of serum
specific antibody levels in RP patients and evaluate the continuous protective
effect of serum specific antibodies on patients with COVID-19. Finally, we
should differentiate RP patients from relapse ones from convalescent subjects,
for who two distinct prevention and control strategies will be adopted.
Taken together, our findings revealed the clinical features of RP patients
who did not show recurrence of clinical symptoms and abnormal laboratory
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint
tests. However, hyper-sensitive detection methods revealed the existence of
SARS-CoV-2 RNA in RP patient specimens tested to be negative using the
commercial kit. Therefore, it is necessary to develop a more accurate
quantitative assessment of the RNA dynamics and additional discharge criteria
to help physicians make a decision. This study provided valuable empirical
information and clinical evidence support for effective management of
COVID-19 patients during convalescent period. Further study should evaluate
the potential clinical significance and transmission risk of RP patients.
Contributors
ZZ, LL, XT, and JA had the idea and for designed the study. ZZ, LL, XT, JA, XJ and QS
had full access to all data in the study and take responsibility for the integrity of the data
and the accuracy of the data analysis. JY, LW, XC, SQ, NL, QC, FW, JC, YL, LL and ZZ
had roles in the clinical management, patient recruitment, sample preparation and clinical
data collection. TX and CS had roles in the RNA and antibody detection experiments, data
collection and analysis. JA, HY, FQ ,YL and ZZ had roles in statistical analysis. JA, XJ, YW,
FZ, XT, LL, YL and ZZ had roles in data interpretation. JA, XL, YF, XT and ZZ wrote the
manuscript. YW, YF, XT and ZZ contributed to critical revision of the report. All authors
reviewed and approved the final version of the manuscript.
Declaration of interests
We declare no competing interests.
Acknowledgements
We acknowledge the work and contribution of all the health providers from Shenzhen
Third People's Hospital.
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint

Reference
1. https://www.who.int/emergencies/diseases/novel-coronavirus-2019.World Health Organization.
2. WHO characterizes COVID-19 as a pandemic.
https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen
(accessed Mar 14, 2020).
3. Guan W-J, Ni Z-Y, Hu Y, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl
J Med 2020. published on February 28.DOI: 10.1056/NEJMoa2002032.
4. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with
COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020. Published Online March
9.https://doi.org/10.1016/S0140-6736(20)30566-3.
5. Wang M, Wu Q, Xu W-Z, et al. Clinical diagnosis of 8274 samples with 2019-novel coronavirus in
Wuhan. medRxiv 2020.published online Feb 18.DOI:10.1101/2020.02.12.20022327.
6. Chen N-S, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019
novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020. published online
Jan 29. https://doi.org/10.1016/ S0140-6736(20)30211-7.
7. Huang C-L, Wang Y-M, Li X-W, et al. Clinical features of patients infected with 2019 novel
coronavirus in Wuhan, China. Lancet 2020.published online Jan 24.https://doi.org/10.1016/
S0140-6736(20)30183-5.
8. Wang D-W, Hu B, Hu C, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel
Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020.published online Feb 7.
https://doi.org/ 10.1001/jama.
9. Lan L, Xu D, Ye G, et al. Positive RT-PCR Test Results in Patients Recovered From COVID-19.
JAMA 2020. Published online February 27. https://jamanetwork.com/on 02/27/2020.
10. Ling Y, Xu S-B, Lin Y-X, et al. Persistence and clearance of viral RNA in 2019 novel coronavirus
disease rehabilitation patients. Chin Med J (Engl) 2020. DOI:10.1097/CM9.0000000000000774.
11. Qu Y-M, Cong H-Y. Positive result of SARS-Co-2 in sputum from a cured patient with COVID-19.
Travel Medicine and Infectious Disease 2020. https://doi.org/10.1016/j.tmaid.2020.101619.
12. Xing Y-H, Ni W, Wu Q, et al. Prolonged presence of SARS-CoV-2 in feces of pediatric patients
during the convalescent phase. medRxiv 2020.published online March 13.
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint

https://doi.org/10.1101/2020.03.11.20033159.
13. http://www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989.shtml (accessed
Mar 9, 2020).
14. Liu Y, Yang Y, Zhang C, et al. Clinical and biochemical indexes from 2019-nCoV infected patients
linked to viral loads and lung injury. Sci China Life Sci 2020;63(3):364-74.
15. Wei L, Ming S-Q, Zou B, et al. Viral Invasion and Type I Interferon Response Characterize the
Immunophenotypes during Covid-19 Infection. SSRN 2020.published online March 18.
https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3555695.
16. Ling Ni, Fang Ye, Chen M-L, et al. Characterization of anti-viral immunity in recovered individuals
infected by SARS-CoV-2. MedRxiv 2020. published online March 20.
https://doi.org/10.1101/2020.03.17.20036640.
17. Zhao J-J, Yuan Q, Wang H-Y, et al. Antibody responses to SARS-CoV-2 in patients of novel
coronavirus disease 2019. medRxiv 2020. published online March 20.
https://doi.org/10.1101/2020.03.02.20030189.
18. Guo X-Q, Guo Z-M, Duan C-H, et al. Long-Term Persistence of IgG Antibodies in SARS-CoV
Infected Healthcare Workers. medRxiv 2020. published online February 14.
https://doi.org/10.1101/2020.02.12.20021386.
19. Liu W, Fontanet A, Zhang P-H, et al. Two-year prospective study of the humoral immune response
of patients with severe acute respiratory syndrome. J Infect Dis 2006;193(6):792-95.
20. Bao L-L, Deng W, Gao H, et al. Reinfection could not occur in SARS-CoV-2 infected rhesus
macaque.medRxiv 2020. published online March 13,
2020.https://doi.org/10.1101/2020.03.13.990226.
21. Li D, Wang D, Dong J, et al. False-Negative Results of Real-Time Reverse-Transcriptase
Polymerase Chain Reaction for Severe Acute Respiratory Syndrome Coronavirus.Korean J Radiol
2020;21(4):505-08.
22. Xia J-H, Tong J-P, Liu M-Y, et al. Evaluation of coronavirus in tears and conjunctival secretions of
patients with SARS-CoV-2 infection SARS-CoV-2 through conjunctiva. J Med Virol 2020. published
online February 26. https://doi.org/10.1002/jmv.25725.
23. Wölfel R, Corman VM, Guggemos W, et al.Virological assessment of hospitalized cases of
coronavirus disease 2019. medRxiv 2020. published online March 8.
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint

https://doi.org/10.1101/2020.03.05.20030502.
24. Lin C, Xiang J, Yan M-Z, et al. Comparison of throat swabs and sputum specimens for viral nucleic
acid detection in 52 cases of novel coronavirus (SARS-Cov-2) infected pneumonia (COVID-19).
medRxiv 2020. published online February 23. https://doi.org/10.1101/2020.02.21.20026187.
25. Zhang F, Abudayyeh OO, Gootenberg JS. A protocol for detection of COVID-19 using CRISPR
diagnostics(v.20200321).
https://www.broadinstitute.org/files/publications/special/COVID-19%20detection%20(updated).pdf
26. Xiao F, Tang M-W, Zheng X-B, et al. Evidence for gastrointestinal infection of SARS-CoV-2. medRxiv
2020.published online February 20. https://doi.org/10.1101/2020.02.17.20023721.
27. Wu Y, Guo C, Tang L, et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet
Gastroenterol Hepatol 2020. Published Online March 19, 2020.
https://doi.org/10.1016/S2468-1253(20)30083-2.
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint
Figure 1 Serial CT imaging of a representative RP and NRP patient. For
the RP patient, the first chest CT scan on admission (day 7 since the onset of
illness) showed ground-glass opacity in both lungs. At 3 day, 6 day and 10 day
after admission (day 10, day 13 and day 17 since the onset of illness), the lung
lesions on the chest CT imaging was significantly reduced accompanied by the
disappeared clinical symptoms. The patient was discharged at day 12 after
admission (day 19 since the onset of illness). At day 26 (day 33 since the onset
of illness), the patient was re-admitted without fever and cough due to positive
RNA detection. The chest CT showed no inflammatory lesions. For the NRP
patient, a chest CT scan showed a small ground glass in the upper left lung on
admission (day 3 since the onset of illness). On day 2 and 8 after admission
(day 5 and day 11 since the onset of illness), the double lower lung lesions
increased significantly on chest CT imaging although the body temperature
and the oxygenation index was returned to normal levels. On day 9, 14 and 17
after admission (day 12, day 17 and day 20 since the onset of illness), the
lesions in both lower lungs were recovered on chest CT imaging. Then the
patient was discharged without fever and cough at day 18 after admission (day
21 since the onset of illness) when SRAS-CoV-2 RNA was also detected to be
negative.
Figure 2 The number of discharge patients and RP patients each day
from Jan 23 to March 10, 2020. On Feb 22, 2020, the anal swab negative test
was added to discharge criterion. Blue, the number of discharge patients. Red,
the number of RP patients.
Research in context
Evidence before this study
Positive SARS-CoV-2 RNA test in some recovered patients has been reported, whose
management has attracted wide attention. However, the number of recovered patients
with the positive SARS-CoV-2 RNA test reported in the literature was small, and the
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint
duration of follow-up was short. The clinical characteristics are lacking and the
potential impact and significance of the patients remain unknown. The lack of these
data makes it difficult to provide empirical information and evidence support for the
management of patients with COVID-19 in the recovery period.
Added value of this study
First, the young and mild COVID-19 patients are prone to be tested positive for
SARS-CoV-2 after discharge. These patients display fewer symptoms, more sustained
remission of CT imaging and earlier RNA negative-conversion but similar plasma antibody
levels during the hospitalization period as compared to those NRP patients. Second,
upon re-admission, these patients show no obviously clinical symptoms and disease
progression. However, the hyper-sensitive detection method potentially recognized false
negative by the commercial kit.
Implications of all the available evidence
The current evidence strongly supports the effective management of COVID-19
patients during their convalescent phase.
cases recovered from COVID-19 tested positive for SARS-CoV-2 after
discharge (re-detectable positive, RP)
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint

Table 1 Baseline characteristics of enrolled patients with COVID-19
RP, re-detectable positive patients; NRP, non-re-detectable positive patients
Mild (n = 30)
Moderate (n = 212)
RP (n = 11)
NRP (n = 19)
P
value
RP (n = 27)
NRP (n = 185)
P
value
Age, median (IQR)-yr
20 (5-64)
23 (2-63)
0.98
38 (2-60)
48 (1-86)
< 0.01
> 14 and ≤ 60 years old-n.(%)
10 (90.9)
18 (94.7)
0.78
26 (96.3)
139 (75.1)
0.11
≤ 14 years old-n.(%)
4 (36.3)
6 (31.6)
0.57
3 (11.1)
6 (3.24)
0.04
> 60 years old-n.(%)
1 (9.1)
1 (5.3)
0.32
1 (3.7)
46 (24.9)
< 0.001
Gender-n.(%)
Male
4 (36.3)
10 (52.6)
0.08
12 (44.4)
90 (48.6)
0.66
Female
7 (63.7)
9 (47.4)
0.12
15 (55.6)
95 (51.4)
0.69
Comorbidities-n.(%)
1 (9.1)
0 (0)
NA
1 (3.7)
41 (22.2)
< 0.01
History of travel or residence in
Hubei-n.(%)
10 (90.9)
16 (84.2)
0.61
23 (85.2)
152 (82.2)
0.82
Fever--n.(%)
2 (18.2)
7 (36.8)
< 0.01
23 (85.2)
133 (71.9)
0.29
Upper respiratory symptoms-n.(%)
5 (45.5)
2 (10.5)
< 0.01
4 (14.8)
34 (18.4)
0.53
Lower respiratory symptoms-n.(%)
5 (45.5)
7 (36.8)
0.34
14 (51.9)
95 (51.4)
0.96
Digestive tract symptoms-n.(%)
0 (0)
2 (10.5)
NA
3 (11.1)
15 (8.11)
0.50
The lesion range of chest CT-n.(%)
Unilateral
0 (0)
0 (0)
NA
6 (22.2)
36 (19.4)
0.66
Multi-lobe of Bilateral
0 (0)
0 (0)
NA
15 (55.6)
105 (56.7)
0.92
All-lobe of Bilateral
0 (0)
0 (0)
NA
6 (22.2)
44 (23.7)
0.82
Chest CT imaging-n(%)
Transient progression
0 (0)
0 (0)
NA
4 (14.8)
67 (36.2)
< 0.05
Sustained remission
0 (0)
0 (0)
NA
23 (85.2)
113 (61.1)
0.05
Steroids use-n.(%)
0 (0)
0 (0)
NA
4 (14.8)
27 (14.6)
0.97
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint

Table 2 RNA detection in the enrolled patients with COVID-19
Mild (n = 30)
Moderate (n = 212)
RP (n=11)
NRP (n=19)
P value
RP (n=27)
NRP (n=185)
P Value
*Days since the onset of illness to
last RNA negative-conversion
17 (11-22)
15 (8-24)
0.71
18 (9-30)
20 (5-47)
0.17
follow-up deadline (March 10)
40 (33-47)
42 (35-49)
0.15
45 (33-54)
46 (30-72)
0.15
discharge
15 (14-22)
16 (10-23)
0.72
17 (9-29)
18 (7-35)
0.47
Days of RNA negative-conversion
since the onset of illness (n, %)
Between 7 and 14 days
3 (27.3)
8 (42.1)
0.08
6 (22.2)
18 (9.7)
0.03
Between 14 and 21 days
7 (63.6)
8 (42.1)
0.04
11 (40.7)
84 (45.4)
0.61
More than 21days
1 (9.1)
3 (15.8)
0.18
10 (37.3)
83 (44.9)
0.40
*Median (range). RP, re-detectable positive patients; NRP, non-re-detectable positive patients
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint

Table 3 Clinical observation of RP patients at re-admission of hospital
Mild (n = 11)
Moderate (n = 27)
Symptoms
fever
0 (0)
0 (0)
cough
1 (9.1)
5 (18.5)
Chest tightness
0 (0)
2 (7.4)
other
0 (0)
3 (11.1)
Chest CT imaging
Normal
11 (100)
10 (37.0)
Stable or absorb
0 (0)
17 (63.0)
Progression
0 (0)
0 (0)
Laboratory examination
Abnormal lymphocyte count
0 (0)
4 (14.8)
increasing serum IL-6 level
0 (0)
0 (0)
increasing serum CRP level
0 (0)
0 (0)
Treatment
Low flow oxygen
0 (0)
4 (14.8)
Traditional Chinese medicine
3 (27.3)
8 (29.6)
Antiviral therapy
0 (0)
1 (3.7)
Number of contacts with symptoms
0
0
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint

Table 4 The comparison between hyper-sensitivity and common
sensitivity detection in RP and NRP patients
Sample
Dates since the
onset of illness
Sherlock
Commercial
S gene
ORF gene
N gene
ORF1 gene
1
Anal swab
44
-
-
-
-
2
Nasal swab
44
+
-
-
-
3
Anal swab
44
+
-
-
-
4
Nasal swab
44
+
+
-
-
5
Anal swab
37
-
+
+
-
6
Nasal swab
37
+
+
-
-
7
Anal swab
42
+
-
-
-
8
Anal swab
43
-
+
-
-
9
Anal swab
30
+
+
-
-
10
Blood
42
+
-
-
-
11
Blood
43
-
-
-
-
12
Blood
30
+
-
-
-
13
Nasal swab
42
-
-
+
-
14
Nasal swab
43
+
-
-
-
15
Nasal swab
30
+
+
-
-
16
Anal swab
32
+
+
-
-
17
Anal swab
37
+
-
-
-
18
Anal swab
36
+
-
-
-
19
Anal swab
37
+
+
-
-
20
Anal swab
41
+
+
-
-
21
Anal swab
37
-
-
+
+
22
Anal swab
31
+
-
-
-
23
Anal swab
43
+
+
-
-
24
Nasal swab
32
+
-
-
-
total
Positive (%)
18 (75%)
10 (41.6%)
3 (12.5%)
1 (4.2%)
25
Nasal swab
47
-
-
-
-
26
Anal swab
47
-
-
-
-
27
Nasal swab
45
-
-
-
-
28
Nasal swab
40
-
-
-
-
29
Anal swab
54
-
-
-
-
30
Anal swab
49
-
-
-
-
31
Nasal swab
44
-
-
-
-
32
Nasal swab
48
-
-
-
-
total
Positive (%)
8 (0)
8 (0)
8 (0)
8 (0)
Note:1-24, re-detectable positive patients; 25-32: non-re-detectable positive patients.
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint

Figure 1
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint
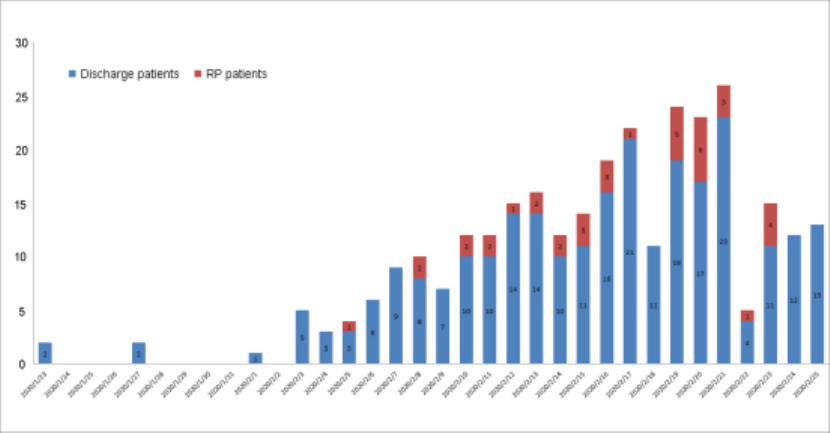
Figure 2
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint

Supplemental Table 1 Analysis of disease severity in RP and NRP patients
Mild (n = 30)
Moderate (n = 212)
Severe (n = 20)
Total (n = 262)
RP
11 (36.7%)
27 (12.7%)
0 (0)
38 (14.5%)
NRP
19 (63.3%)
185 (87.3%)
20 (100%)
224 (85.5%)
RP, re-detectable positive patients; NRP, non-re-detectable positive patients
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint

Supplemental Table 2 Analysis of serum anti- SARS-CoV-2 IgG and IgM antibody
levels in patients with covid-19 at discharge
Mild
Moderate
RP (n = 10)
NRP (n = 10)
P value
RP (n = 21)
NRP (n = 119)
P value
Serum IgG (n, %)
High
1 (10%)
0 (0)
0.30
8 (38.1%)
41 (34.5%)
0.95
Medium
6 (60%)
4 (40.0%)
10 (47.6%)
60 (50.4%)
Low
3 (30%)
6 (60.0%)
3 (14.3%)
18 (15.1%)
Serum IgM (n, %)
High
1 (10%)
0 (0)
0.59
7 (33.3%)
39 (32.8%)
0.68
Medium
7 (70%)
8 (80.0%)
12 (57.1%)
60 (50.4%)
Low
2 20%)
2 (20.0%)
2 (9.5%)
20 (16.8%)
Note: RP, re-detectable positive patients; NRP, non-re-detectable positive patients
High, titer with more than 16400; Medium, titer between 5400 and 16400; Low, titer with less than 5400
All rights reserved. No reuse allowed without permission.
perpetuity.
preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in
The copyright holder for thisthis version posted March 30, 2020. ; https://doi.org/10.1101/2020.03.26.20044222doi: medRxiv preprint
